We will have posterboards with dimensions of 48" by 48". The recommended poster size is 36" by 48" (either portrait or landscape). A larger poster with a size of 48" by 48" can be accommodated, and it should be fine if you exceed this limit by an inch or so.
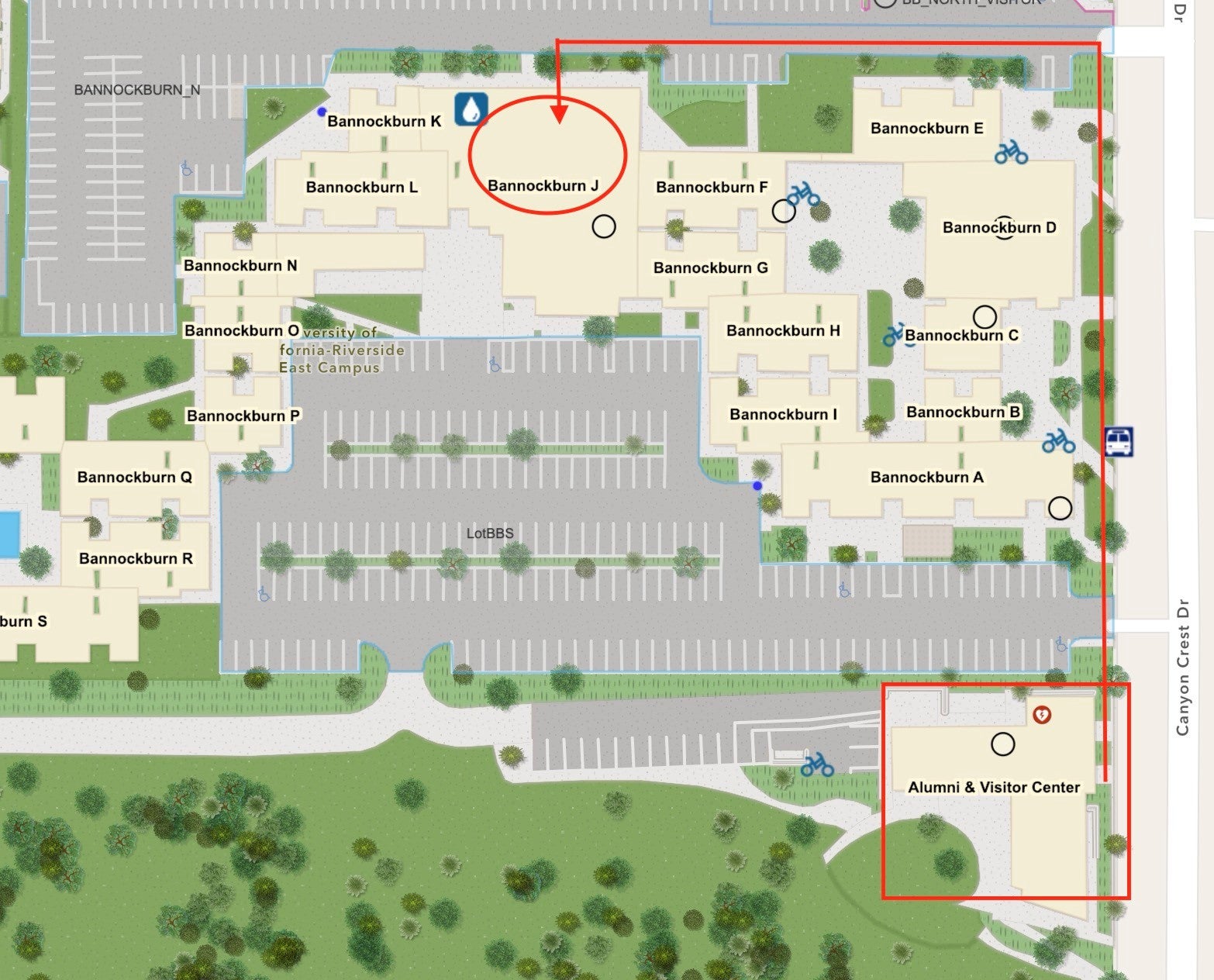
The Poster session will be in Bannockburn J102, located in Bannockburn Village. This meeting room is less than a 5-minute walk from the Alumni and Visitor's Center. Please reference the image below.
Posters for Friday the 17th
Andrew Willems - Poster # 1
University of Tennesse
Genome Science & Technology
Graduate Student
Title: A Gene-Expression-Based Model for Improving SCLC Survival Prediction and Biomarker Discovery
Abstract: Small cell lung cancer (SCLC) is an aggressive and recalcitrate type of lung cancer, posing a significant global health challenge with substantial morbidity. SCLC is characterized by its rapid metastasis, low survival rates, and unique histopathological features. There is therefore a critical need for novel biomarkers that can enhance patient survival prediction. In this work, we introduce an expression-based model that leverages single-cell RNA sequencing data and miRNA targeting information. Our preliminary results show satisfactory accuracy of SCLC patient survival prediction. Because of the model’s interpretability and lightweight nature, it is feasible to transfer the trained model across datasets. Additionally, we show that our model outperforms commonly used biomarker discovery methods, underlining its potential to enhance SCLC prognosis while also providing tangible biological insights. awillems@vols.utk.edu
Anthony Zamora - Poster # 2
UCI Mathematics
Graduate Student
Title: Spatial Stochastic Modeling of Evolutionary Processes: Applications to Cancer and Biofilms
Abstract: In this project, we study patterns of evolutionary and ecological dynamics in spatial systems, and consider two separate applications. (1) Melanoma is a widespread cancer, comprising 1.7\% of all new cancer diagnoses and about 70\% of all skin cancer-related deaths. Melanoma is treatable by BRAF inhibitors that directly interfere with the processes that govern cell growth and signaling to destroy susceptible cancer cells. However, research has shown that treatment with such inhibitors can become complicated as cell resistance develops in the patient. These resistant cells form such a strong dependence on treatment doses that they become advantageous in the presence of a BRAF inhibitor but lose this advantage in its absence. We investigate the dynamics of susceptible and resistant cells under the administration of intermittent treatment, by first analyzing an ODE model, and then developing spatial stochastic models. Analysis thus far has yielded conditions for elimination of both susceptible and resistant cell populations as well as an optimal strategy for intermittent treatment when both periods of on and off treatment are constant. The spatial model describes a more realistic system and can simulate elimination of cancer under a wider range of kinetic parameters. (2) We also use the spatial stochastic model to study the dynamics of thin layers of bacteria called biofilms. More specifically, we consider the bacteria Vibrio Cholerae which form 2D colonies on the surface of small aquatic crustaceans in the ocean. These bacteria release polysaccharides and proteins to form a substance that allows them to attach to surfaces and surrounding bacteria. There are however bacteria that do not produce the substance, which allows them to gain advantage in reproduction. We call individuals that produce this substance Producers (P), and those that do not Lazy (L) cells. P cells, although they reproduce slower, gain advantage over L cells under the high flow conditions, where they are washed out to a lesser extent compared to P cells. The spatial stochastic model is modified to investigate the competition dynamics of P and L cells, and to study their spatial patterning, cluster size and density of bacteria, as relevant parameters are varied. zamoraa3@uci.edu
Audrey Oliver - Poster # 3
San Diego State
Department of Mathematics and Statistics
Computational Science Research Center
Graduate Student
Title: Modeling the Spatiotemporal Distribution of HIV Infection in the Brain
Abstract: In standard clinical practice, only plasma viral load and CD4 counts are measured to keep track of disease status and progression in Human Immunodeficiency Virus (HIV)-infected individuals. However, viruses reside in the brain, causing neurocognitive disorders and an obstacle to a cure, despite virus control in plasma with antiretroviral therapy. Therefore, tracking the virus distribution across different brain compartments is essential for disease management in HIV-infected individuals. In this study, we first performed a correlation network analysis of RNA in the brain with plasma and CSF (Cerebrospinal fluid) to identify whether plasma or CSF viral loads can infer the viral burden in the brain. Secondly, we performed a correlation network analysis of viral RNA among different brain regions to identify the brain’s essential regions related to viral burden within the brain. Thirdly, we built a mathematical model that explains the spatiotemporal distribution of HIV in the brain using the essential brain regions obtained from our correlation analysis. Our model was validated using data collected from the brain of the simian immunodeficiency virus (SIV)-infected macaques. We analyzed the model and performed parameter sensitivity to get insights into the distribution and replication of HIV throughout the different brain regions, as well as evaluate the reproduction number to determine the persistence of the virus. akoliver@sdsu.edu
Austin Hansen - Poster # 4
University of California, Riverside
Mathematics
Graduate Student
Title: Computational Model of Pseudomonas fluorescens Migration in Response to L. bicolor
Abstract: Mitigating climate change requires large-scale shifts towards alternative energy sources. One such source is biofuels, which face a key challenge preventing them from being adopted: fertile land is a finite source facing competition between food and biocrops. Root colonization by arbuscular mycorrhizal fungi and helper bacteria has been shown to increase plants’ ability to thrive in less nutrient-dense environments. However the fundamental interactions between bacteria and fungi which allow the formation of this symbiotic relationship are still largely unknown. In order to illuminate some of the mechanistic factors that can further promote this growth, we build a computational model of bacterial migration. The model is then calibrated to 3 separate bacteria including Pseudomonas fluorescens, P. Aeruginosa, and P. Putida. Simulations are carried out in both a liquid environment as well as in the presence of a simple fungal network. Preliminary results suggest that both the strength of chemical gradient as well as the angle change between bacterial swimming modes play an important role in their ability to find and distribute along the fungi. ahans016@ucr.edu
Daisy Ulloa - Poster # 5
San Diego State
Computational Science Research Center
Graduate Student
Title: External tensional force applied to mice mammary glands alters the length and branching angles of the ductal network
Abstract: The mammary gland is one of the structures in the body to undergo most of its development postnatally. During puberty, the mammary gland forms an extensive network of epithelial ducts that are vital for milk secretion and breastfeeding. Mechanical forces are believed to play a key role in the development of the ductal network; however, this process is not fully understood. We specifically aim to understand if external mechanical forces impact pubertal branching morphogenesis and the interplay between external and internal forces during ductal formation. At five weeks, the skin surrounding the left abdominal number four mammary gland nipple of female mice were adhered together with surgical glue to create a tensional force (TEN). After two weeks (seven weeks of age), the abdominal glands were removed and whole mounts prepared. Using image analysis, we compared the ductal morphology of the TEN glands to the left abdominal number four glands from untreated-control (CTL) mice. We found that the TEN glands grew significantly longer than the CTL glands, despite no significant difference in size. Upon further analysis, we found significance in the branching angle between TEN and CTL. In-silico simulation of branching morphogenesis show branching angles create a significant difference in the length between TEN and CTL glands. These results imply external tensional forces alter the length of the overall gland. These results show a need to further understand how the mechanical environment impacts mammary gland development, as well as other organs, and their long-term functions. dulloa3712@sdsu.edu
David Frankhouser - Poster # 6
City of Hope
Computational and Quantitative Medicine
Faculty
Title: State-transition model of chronic myeloid leukemia predicts disease evolution and treatment response
Abstract: Chronic myeloid leukemia (CML) is a well-studied hematological malignancy driven by the expression of the BCR-ABL fusion gene. Current treatments using tyrosine kinase inhibitors (TKIs) are highly effective for CML, but lack of compliance, drug intollerance or BCR/ABL mutations may favor disease relapse or progression to blast crisis in some patients. To better understand the dynamics of disease initiation, growth, and response to therapy, we applied a state-transition mathematical approach. Using a mouse model of CML that has a tetracycline-off inducible BCR-ABL gene, we performed bulk RNA-seq from time-series blood samples. We constructed a CML state-space of the transcriptome and showed that it undergoes state-transition during CML development moving in a state-space characterized by a leukemogenic potential. Using vector analysis of differentially expressed genes (DEG) and gene set enrichment analysis (GSEA), we showed distinct anti-CML processes at the early critical point that opposed leukemic transformation, followed by a progressively expanding CML-promoting transcriptional program that included, among others, upregulated inflammation, angiogenesis and metabolism-related processes. To investigate therapeutic response, we treated the mice either with nilotinib or suppressed the otherwise Tet-off inducible BCR-ABL expression with re-administration of tetracycline [called teff-off/te-on (TOTO)]. When the two treatment approaches were incorporated to our state-transition model, we observed dramatically different effects. Although TKI reduced the circulating myeloid population, the state-transition model showed that the transcriptome still represented a diseased state, and in fact, these mice developed CML immediately after therapy was concluded. Conversely, turning off BCR-ABL expression with TOTO returned transcriptome to a perturbed healthy state, suggesting that once BCR-ABL transformation had occurred, the transcriptomes would not return to a completely healthy state even if BCR/ABL is completely suppressed. These results support the applicability of state-transition analysis as a valuable approach to gain real time insights into disease development, progression and ongoing treatment response. dfrankhouser@coh.org
Ghizelle Abarro - Poster # 7
University of Tennessee, Knoxville|
Chemical and Biomolecular Engineering
Graduate Student
Title: Novel, Model-derived Hypotheses on the Regulation of BRUTUS Activity in Plants’ Iron (Fe) Homeostasis
Abstract: Iron (Fe) is a critical nutrient, and we depend on plants to act as our conduit of Fe from the earth. As plants assimilate Fe as they grow, they function constantly at the edge of iron deficiency. In response to iron deficiency, plants activate a complex system of signaling events to increase Fe uptake. However, Fe is a potent redox agent, and its excess causes oxidative damage. Thus, Fe deficiency responses must be regulated to ensure nutrient availability without exposing cells to a potent cytotoxin. A key player in the deficiency response of Arabidopsis Thaliana is BRUTUS (BTS), an Fe-binding enzyme responsible for ubiquitinating proteins involved in Fe handling. BTS knock-down mutants accumulate more Fe, suggesting that BTS limits Fe uptake. However, wild-type plants respond to Fe deficiency by upregulating BTS – suggesting that BTS might be implicated in activating Fe uptake. Increased Fe has been observed to destabilize BTS – again supporting the notion that BTS should be active under deficiency and inactive when Fe is abundant. It is unclear why plants lacking a key deficiency response protein would exhibit robustness to Fe deficiency. And if BTS acts as a “brake” on Fe uptake, why upregulate BTS precisely when Fe is needed? Herein, we present a study of the Fe-dependent activity of BTS as viewed through the lens of a mathematical model. Embedding known subcellular events and their putative Fe-dependence into a system of differential equations, we generated simulations predicting cytosolic and nuclear accumulation of BTS, as a function of cytosolic Fe. We then used simulation-based inference to determine what kinetics would be consistent with known emergent behaviors of this cellular system. In doing so, we identified a new role for Fe in regulating BTS and thereby posited a non-monotonic relationship between cytoplasmic Fe and the BTS “brake”. gabarro@vols.utk.edu
Hyunah Lim - Poster # 8
University of Maryland, College Park
Mathematics
Graduate Student
Title: Mathematical modelling of the impacts of screening and vaccination in HPV
Abstract: Cervical cancer is one of the most frequent cancers women may suffer, which caused about 342,000 death cases in 2020 worldwide. It is known that cervical cancer is highly attributable to HPV, while HPV has no medical treatment leading to cure of infection but realistic prevention only available from vaccine. However, HPV vaccines are overly expensive for individuals, and many countries including South Korea has been vaccinating young female population only for cost-effectiveness. We wish to study the impacts of different vaccine strategies such as vaccinating males and females altogether in South Korea, and also experiment on the impact of detection in early cancer stages for females through screening, to find the ideal strategy for our society to take. Although we experiment on South Korean data, our model is not regional, and we expect it to contribute to researches similar in different countries also. hyuna913@umd.edu
Isaac Tate - Poster # 9
UC Riverside
Mathematics
Postdoctoral Fellow
Title: A coupled growth and signalling model for Shoot Apical Meristem in Arabidopsis
Abstract: In a project studying the classification of stem cells in Arabidopsis, I am studying the chemical signaling of the transcription factors in the shoot apical meristems. The regulation of the homeodomain transcription factor WUSCHEL concentration is critical for stem cell homeostasis in Arabidopsis shoot apical meristems. Transcription factors to note are WUSCHEL, which regulates the transcription of CLAVATA3 through a concentration-dependent activation-repression switch, and CLAVATA3, a secreted peptide that activates receptor kinase signaling to repress WUSCHEL transcription. itate001@ucr.edu
Jennifer Rangel Ambriz - Poster # 10
University of California, Riverside
Mathematics
Graduate Student
Title: Combined Modeling and Experimental Study of Shape Formation of the Drosophila Wing Imaginal Disc
Abstract: Developmental mechanisms of tissue growth and shape formation are not well understood. Combined experiments and multi-scale computational model of the Drosophila wing imaginal disc will be used to show that nonhomogeneous actomyosin patterning defines the local basal curvature and nuclear positioning while cell proliferation enhances the local basal curvature. A coarse-grained stochastic model of actomyosin dynamics will be described to capture the directionality of actomyosin contractile forces and nuclear positioning during cell growth and division. jrang008@ucr.edu
Konstancja Urbaniak - Poster # 11
City of Hope
Quantitative and Qualitative Medicine
Postdoctoral Fellow
Title: Deciphering the Interplay of IL6 and IL10 in Immune Signaling Networks in Breast Cancer Patients vs. Healthy Donors Through Top-Down Network-Centered Analysis
Abstract: Interleukins (ILs) are vital mediators of immune response and cancer progression, with IL6 and IL10 being of particular interest in Breast Cancer (BC). IL6 signaling has been associated with worse patient prognosis, while multifaceted effects of IL10 enhance tumor survival and metastasis. Despite their significance, IL6 and IL10 have not been simultaneously studied in BC immune system. One of the reasons is a lack of interpretable network-centered analytical framework for complex multivariable analyses. In this study, we aimed to develop experimental-computational methods for investigating complex effects of cytokine combinations in health and disease, with an example application of dissecting IL6 and IL10 treatment of PBMC samples from 10 healthy donors and 17 BC patients. We performed 18-channel FACS to explore proteins of immune cell surface receptors and the JAK-STAT signaling pathway. We employed Bayesian network modeling using our custom BNOmics software to assess how immune signaling networks are re-wired under different conditions. Evaluating over 500 networks highlighted differential patterns of dependencies in the immune signaling networks in BC patients vs healthy donors, with and without cytokine stimulation. We identified key fulcrum points in the immune signaling network, revealing established and novel modulators in BC. Our fully data-driven, high-dimensional analysis pipeline can be seamlessly applied to any network comparison scenarios, offering high interpretability leading to novel hypothesis generation and biological insight. kurbaniak@coh.org
Kwadwo (Kojo) Bonsu - Poster # 12
University of California, Irvine
Chemical & Biomolecular Engineering
Graduate Student
Title: Mathmetical Modeling & Bioinformatics Analysis to Investigate Multigenerational Epigenetic Stability of DNA Methylation Landscapes
Abstract: Currently, it is not well understood what factors contribute to the stability of the epigenome in different cell types across the lifetime of mammals. Better understanding of these factors would aid development of therapies for age-related diseases, including cancer. DNA methylation is a widely studied epigenetic mark that is located primarily on Cytosine-phosphate-Guanine (CpG) dinucleotides and is associated with transcriptional gene silencing. CpG Islands (CGIs) are regions of the DNA which contain high CpG content. Bioinformatic analysis of methylation landscapes in various human cell lines shows that methylation levels of CGIs are inversely correlated with CpG Island size, and suggest a size threshold exists, below which islands are more likely to be methylated. In contrast, preliminary analysis of mouse cell lines shows that the methylation levels of CGIs are not strongly dependent on island size. These observations from the data are used to inform and constrain stochastic mathematical models of CpG methylation dynamics. The simulations predict that the size and density of CGIs influence overall stability of their methylation patterns, reflected in variable state-switching of island methyl states across multiple replication cycles. However, the stability is also strongly dependent on the strength of interaction between neighboring CpGs in (de-) methylating reactions. Together, the stochastic models and bioinformatic analysis suggest that (de-)methylating enzymes impose different CpG-interaction strengths and lengthscales in mouse versus human, with implications for stability of the epigenetic landscape." kbonsu@uci.edu
Maxim Kuznetsov - Poster # 13
City of Hope
Department of Computational and Quantitative Medicine
Postdoctoral Fellow
Title: Mathematical Modeling Unveils Optimization Strategies for Targeted Radionuclide Therapy of Blood Cancers
Abstract: The demand for selective anticancer treatments gave rise to targeted therapy. A relevant technique is conjugation of cancer-specific antibodies with radioactive isotopes, or nuclides, with their subsequent intravenous injection. Targeted radionuclide therapy (TRT) is not devoid of side-effects, and its toxicity is a more complicated issue compared to that of external beam radiotherapy. We have developed a mathematical model based on our experiments on disseminated multiple myeloma mouse model and its treatment by 225Ac-DOTA-daratumumab, specific to CD38 receptors overexpressed in these cancer cells. This model was investigated with the key goal of suggesting methods to increase efficacy and/or reduce toxicity of single-dose and multidose treatments. It yielded the following conclusions. 1) The ratio of labeling of nuclides to antibodies plays a crucial role in determining the treatment efficacy and toxicity. Large labeling ratio and/or drug dilution with unlabeled antibodies can compromise achievement of sufficient curative density of radionuclides on the cancer cells, due to their excessive occupation by unlabeled antibodies.
2) Proper dosing can be performed via estimation of the total binding capacity of cancer cells. In blood cancer setting this can be achieved via preliminary injection of a small test dose and measuring the initial pharmacokinetic curve of the radiolabeled antibodies. The following dose, used for curative intent, should not significantly exceed the cancer capacity, as the nuclides freely floating in blood would have negligible efficacy-to-toxicity ratio.
3) The efficacy of the treatment strongly correlates with the dose delivered to the yet viable cancer cells, as opposed to the cells which are already lethally damaged by radiation. With a single potentially curative dose, only a small fraction of radioactivity (<4%) can be spent on viable cells. At that, newly injected antibodies will distribute passively between the receptors of viable and doomed cells. The key to the increase of TRT efficacy in multidose setting is the increase of exposure of yet undamaged cancer cells to newly injected doses of radioconjugates. There are two ways to achieve it. 4) Under broad assumptions, increase of multidose TRT efficacy can be obtained by proper timing of doses, adjusted for the moments when sufficient number of damaged cancer cells is eliminated from the system, while regrowth of viable cells is yet moderate. 5) In the considered 225Ac-DOTA-daratumumab setting the following assumptions should be valid: alpha-particles have short range; binding affinity is high, and receptors are quickly internalized; unlabeled antibodies can be injected in great amounts without significant toxic effects. Under these circumstances, the increase of multidose TRT efficacy can be achieved by muting damaged cells with unlabeled antibody after the first dose of radioconjugates, so that the further injected radioconjugates will be effectively redirected to yet viable cells, which continue to proliferate and to produce specific receptors. 6) In real-life conditions, when the estimation of corresponding model parameters is limited or impossible, repetitive injections of radioconjugates diluted with unlabeled antibodies, with caution taken for not exceeding the toxicity limits and not exceeding the current binding capacity of cancer cells, should automatically take advantage of the above described methods for increasing TRT efficacy." mkuznetsov@coh.org
Mayesha Sahir Mim - Poster # 14
University of Notre Dame
Chemical and Biomolecular Engineering
Graduate Student
Title: Balancing competing effects of tissue growth and cytoskeletal regulation during Drosophila wing disc development
Abstract: How a developing organ robustly coordinates the cellular mechanics and growth to reach a final size and shape remains poorly understood. Through iterations between experiments and new model simulations that include a mechanistic description of interkinetic nuclear migration, we show that the local curvature, height, and nuclear positioning of cells in the Drosophila wing imaginal disc are defined by the concurrent patterning of actomyosin contractility, cell-ECM adhesion, ECM stiffness and interfacial membrane tension. We show that increasing cell proliferation via different growth promoting pathways results in two distinct phenotypes. Triggering proliferation through insulin signaling increases basal curvature but an increase in growth through Dpp signaling and Myc causes tissue flattening. These distinct phenotypic outcomes arise from differences in how each growth pathway regulates contractility, cell-ECM adhesion and ECM stiffness. The coupled regulation of proliferation and cytoskeletal regulators is a general strategy to meet the multiple context-dependent criteria defining tissue morphogenesis." mmim@nd.edu
Mustafa Farahat - Poster # 15
University of Tennessee, Knoxville
Department of Chemical and Biomolecular Engineering
Graduate Student
Title: Mechanistic Modelling of DNA Damage Repair Process
Abstract: Impairment of the DNA damage response (DDR) is a characteristic of nearly all cancers. Fortunately, the associated genetic defects in cancer can be exploited in targeted anticancer therapies that induce context-specific lethal outcomes for cells bearing DDR defects. However, several challenges remain to realize the promise of these therapies. For example, healthy cells may be compromised, yielding side effects and toxicity. Aside from toxicity, which is a common challenge in drug discovery, the emergence of therapeutic resistance is currently the leading cause of unsuccessful cancer treatment. To clarify the context-specific effects of DDR-targeted interventions, we are developing an integrated stochastic model to elucidate the complex intercellular mechanism of DNA damage repair in cancer. The model will deepen our understanding of the key determinants of competition between DDR pathways, their perturbation in cancer cells, and their evolution in response to resistance-driving mutations. The model will also predict the consequent cell fate in response to radiation-induced DNA damage and how this fate varies as a function of radiation dose and cell cycle phase during exposure. mfarahat@vols.utk.edu
Patrick Lawton - Poster # 16
University of California, Riverside
Biophysics
Graduate Student
Title: Interspecific dispersal influences pattern formation in model metacommunities
Abstract: Organisms exhibit diverse behavioral responses to environmental conditions, with dispersal behaviors standing out as a significant driver of ecosystem dynamics across spatial scales. Different species follow unique sets of behavioral rules to decide whether to leave a habitat, leading to variation in resource tracking, competitor avoidance, and predator evasion in space among taxa. Understanding the impact of interspecific interactions on dispersal is therefore vital for linking species interaction dynamics with large scale spatial patterns in community structure. Modern metacommunity models often simplify dispersal as a random diffusion process with constant \emph{per capita} rate, overlooking the influence of interactions with other species on animals' decisions to leave a habitat. Here, we show how inclusion of interspecific dispersal rules influence the emergence of heterogeneous spatial patterns in metacommunities. We first focus on common three-species interaction motifs, in each case computing master stability functions that separate the contributions of local and spatial interactions on metacommunity dynamics. Our analysis reveals that the inclusion of interspecific dispersal terms generally promotes spatial pattern formation across each interaction motif, resulting in the emergence of either static spatial patterns or traveling spatial waves. However, when ecologically reasonable constraints are imposed on the dispersal response, reflecting adaptive behaviors such as resource tracking and predator avoidance, a strong homogenizing effect is observed. We generalize our findings to species-rich, random ecological metacommunities to show that both unconstrained and constrained interspecific dispersal rules have qualitatively similar effects on spatial patter formation to those observed in three species motifs. plawt001@ucr.edu
Rachel Hobbs - Poster # 17
San Diego State University
Math and Statistics
Graduate Student
Title: Differential protein expression analysis of point mutations to IDH1
Abstract: Point mutations to isocitrate dehydrogenase 1 (IDH1), a homodimeric enzyme, is a well-known driver of the vast majority of lower grade gliomas and secondary glioblastomas. Both mutant and wild type (WT) IDH1 have been implicated in cancer, and mutant IDH1 is a target for therapies. In this study, our primary objective was to understand the proteome differences between the extracellular vesicles of IDH1 WT and three mutants, R132Q, R132H, and R132H /+ KnockIn, using mass spectrometry-based proteomics. We conducted differential protein expression analysis and overrepresentation analysis to identify the unique protein signatures and functional pathways associated with each extracellular vesicles of IDH1 variant utilizing statistical models. Preliminary findings show that a total of 187 proteins were differentially expressed between R132Q vs. WT; 159 proteins between R132H vs WT; 503 proteins between R132H/+ KnockIn vs WT; and 167 proteins between R132Q vs. R132H. A total of 0 pathways for R132Q vs WT; 104 pathways for R132H vs WT; 144 pathways for R132H/+ KnockIn vs WT; and 0 pathways for R132Q vs. R132H were enriched with these proteins. This analysis shows the potential of proteomics and other -omics technologies in understanding IDH1’s role in cancer development. Future work will focus on understanding how the enriched pathways affect tumor suppression or growth. rhobbs8312@sdsu.edu
Rachel Sousa - Poster # 18
UC Irvine
Mathematical, Computational, and Systems Biology
Graduate Student
Title: Identifying Critical Immunological Features of Tumor Control and Escape Using Mathematical Modeling
Abstract: The immune system can eradicate cancer, but various immunosuppressive mechanisms active within a tumor curb this beneficial response. Cytotoxic T cells (CD8s), regulatory T cells (Tregs), and antigen-presenting dendritic cells (DCs) play an important role in the immune response; however, it is very cumbersome to unravel the effects of multimodal interactions between tumor and immune cells and their contributions to tumor control using an experimental approach. Thus, we leverage the power of mathematical modeling to identify critical immunological features associated with tumor control and escape. We developed a mathematical model of CD8s, Tregs, and DCs and used an automated model selection procedure to identify regulatory feedback mechanisms sufficient to reproduce experimentally observed behaviors that regulate the immune system. After determining stable regions of parameter space, we included tumor cells in the model. This tumor-immune model captures the influence of tumor immunogenicity on tumor control and escape, the role of PD-L1 in tumor progression, and the key role of CD8s in rejecting the tumor and of Tregs in promoting tumor acceptance. We use the model to explore open questions in tumor immunology. Namely, we investigate what mechanisms facilitate negative feedback between Tregs and CD8s, how important cytokines are at prolonging T cell lifespan, and optimal immunotherapy scheduling to stimulate CD8 but not Treg activation. This model will accelerate tumor immunotherapy research by allowing for rapid testing of any combination of therapies and schedules that can then be validated experimentally, while also being able to explore mechanisms not yet testable in vivo. rssousa@uci.edu
Reagan Hsu - Poster # 19
University of California, Irvine
Mathematics
Research Intern
Title: ANOMALOUS ULTRASENSITIVE COMPOSITE REACTIONS IN CELLULAR SIGNALING
Abstract: Ultrasensitivity is a crucial cellular mechanism that plays a vital role in signal transduction. Cells employ ultrasensitivity through "switches" that respond to small changes in signaling factor concentrations. Such switches exhibit steep sigmoidal behavior and are characterized by the Hill coefficient. Traditionally, pairwise combinations of these switches were thought to adhere to Ferrell's inequality, which sets limits on their composed ultrasensitivity. However, recent research has disproven Ferrell's inequality, suggesting the existence of biological systems that can combine non-ultrasensitive reactions to achieve high ultrasensitivity. The specific class of reactions that accomplish this optimization remains unknown. In this paper, we utilize mathematical models to explore how variations in molecule saturation as well as reaction gradient impact ultrasensitivity. These variations result in differences in concavity, steady-state values, and composition structure of the functions. Our findings demonstrate that combining robust functions with threshold curves disprove Ferrell's inequality, generating exceptionally high levels of ultrasensitivity. This discovery could provide novel insights into understanding physiological systems that exhibit switch-like behavior and contribute to the defining of a broader set of reactions for which Ferrell's inequality does not hold true. Here, we specifically discuss the composition of two systems: multisite protein binding and absolute robustness. reaganhsu123@gmail.com
Tyler Simmons - Poster # 20
University of Maryland
Institute for Physical Science and Technology
Graduate Student
Title: Modeling the Tumor-Immune Stalemate During the Development of T cell Exhaustion
Abstract: Consistent tumor burden results in an overexposure and overstimulation of antigen specific T cells. As a result of this overstimulation, T cells will become dysfunctional. This form of dysfunction is characterized by diminished effector functions, a state of hypo-functionality referred to as exhaustion. Exhausted CD8+ cytotoxic T cells continue to combat the tumor burden but are ultimately unsuccessful, resulting in a tumor-immune stalemate. Although the mechanisms behind cellular exhaustion have not been fully elucidated, targeting exhausted T cells to restore effector functions has shown to be a promising form of immunotherapy. Presented here is a mathematical framework that highlights the development of exhaustion in CD8+ T cells and the formation of the tumor-immune stalemate. Analysis of this model identifies key aspects of this system to examine and prioritize for future immunotherapy strategies. tsimms16@umd.edu
Widodo Samyono - Poster # 21
Jarvis Christian University Sciences
Mathematics and Engineering
Faculty
Title: Methods for solving PDE-constrained optimization in modeling cancer growth predictions and treatment responses
Abstract: This presentation discusses what underrepresented undergraduate students at Jarvis Christian University, Hawkins, Texas, conducted in their research for in vitro experiments for inducing nanoparticles into cancer cells. Additionally, they have been exploring the methods for solving PDE-Constrained Optimization as the mathematical models for the data from the biology laboratory experiments. To find the best fit model to the data, they used and compared the classical mathematical models for cancer growths and treatments and set up the problems as unconstrained optimizations. wsamyono@jarvis.edu
Haleh Alimohamadi - Poster # 22
UCLA Bioengineering
Postdoctoral Fellow
Title: LL37 in skin inflammation and cardiovascular risk: unraveling the connection between autoimmune diseases and atherosclerosis through mechanistic and machine learning insights
Abstract: LL37 is a human cathelicidin innate immune peptide that is implicated in the pathogenesis of autoimmune diseases such as lupus, rosacea, and psoriasis. Recent studies have highlighted a significantly increased risk of cardiovascular disease in patients with psoriasis and other inflammatory diseases. Particularly, the severity of psoriasis has been positively correlated with an elevated likelihood of developing atherosclerosis. However, the mechanochemical mechanism underlying the association between inflammatory skin diseases and the increased risk of atherosclerosis remains elusive. In this study, we investigated the role of LL37 in promoting Low-Density Lipoprotein (LDL) uptake and lipid accumulation in macrophages and endothelial cells. We found that LL37 enhances macrophage LDL uptake through receptor-mediated endocytosis. Using high-resolution Small Angle X-ray Scattering (SAXS) experiments, we showed that the interaction between LDL particles and LL37 leads to a significant increase in the LDL size. This enlargement provides a larger surface area for receptor binding and also reduces the energy required for membrane bending during endocytosis. Notably, our results demonstrated that LL37-induced LDL uptake is unique to humans and primates within the cathelicidin antimicrobial gene family and is not observed in mice or rabbits. In vivo studies using humanized LL37-transgenic mice on a high-fat diet demonstrate that these mice develop larger atheroma plaques than control mice. Developing a novel generative and unsupervised machine learning framework, we compared the correlative physiochemical distance between cathelicidin peptides from various species and a large data set of curvature-generating peptides. This enabled us to identify LL37-like fragments e.g., in viral infections such as SARS-CoV2 that can be correlated with long-term cardiovascular sequelae in patients who suffered from severe Covid19. halimohamadi@ucla.edu
Yi Fu - Poster # 23
University of California, San Diego
Bioinformatics and Systems Biology
Graduate Student
Title: Stochastic Analysis of Chromatin Modification Circuits that Control Epigenetic Cell Memory
Abstract: Epigenetic cell memory, the inheritance of gene expression patterns across subsequent cell divisions, is a critical property of multi-cellular organisms. It was previously found via simulations of stochastic models that the time scale separation between establishment (fast) and erasure (slow) of chromatin modifications (such as DNA methylation and histone modifications) extends the duration of cell memory, and that different asymmetries between erasure rates of chromatin modifications can lead to different gene expression patterns [1]. We provide a mathematical framework to rigorously validate and extend beyond these computational findings [3]. These stochastic models can be viewed as examples of Stochastic Chemical Reaction Networks (SCRNs). For our study of epigenetic cell memory, these are singularly perturbed, finite state, continuous time Markov chains. We exploit certain special structure in our models and extend beyond existing theory, to determine the behavior of stationary distributions and mean first passage times between states for these singularly perturbed Markov chains when the perturbation parameter is small. In particular, we provide an algorithm to determine the orders of the poles of mean first passage times and the orders of leading entries in the stationary distribution, and we also characterize the leading coefficients in the series expansions for the stationary distribution and mean first passage times in terms of a reduced Markov chain. Then, we focus on determining how different erasure rates for chromatin modifications affect the behavior of our chromatin modification circuit models. This study is conducted by exploiting comparison theorems for Markov chains that we recently developed in [2]. We also determine analytical expressions for upper and lower bounds for the mean first passage times for continuous time Markov chains having a certain independent level structure. The theoretical tools developed in our work not only allow us to set a rigorous mathematical basis for highlighting the effect of chromatin modification dynamics on epigenetic cell memory, but they can also be applied to other singularly perturbed Markov chains beyond the applications in this paper, especially those associated with chemical reaction networks. yif064@ucsd.edu
Reference
[1] Bruno S, Williams RJ, Del Vecchio D. Epigenetic cell memory: The gene’s inner chromatin modification circuit. PLOS Computational Biology 18(4): e1009961 (2022). [2] Campos FA, Bruno S, Fu Y, Comparison Theorems for Stochastic Chemical Reaction Networks. Bull Math Biol 85, 39 (2023).
[3] Bruno S, Campos FA, Fu Y, Del Vecchio D, Williams RJ. Stochastic analysis of chromatin modification circuits that control epigenetic cell memory (submitted)
Navaira Sherwani - Poster # 24
University of California, Riverside
Title: Insights into Bud Morphogenesis Dynamics in Aging Yeast
Abstract: Understanding cellular aging is crucial for extending organismal lifespan and studying age-related degenerative diseases. In budding yeast recent experiments revealed two aging modes—nucleolar and mitochondrial decline. Bud dimensions measured over a cell cycle show linear growth in both modes. In one, cells maintain bud size and spherical shape, whereas bud size increases and shape becomes tubular in the other. We introduce a chemical-mechanical coupled model predicting that linear bud growth results from delivering new cell surface materials to Cdc42 polarization at a constant rate. Simulations confirm the generation of elongated buds by locally inserting materials at the bud tip. These findings suggest cellular aging may impact the maintenance of chemical signaling polarization that directs the delivery of new materials. Our aim now is to simulate aging under varying environmental conditions with cellular signalling and determine the role played by the septin neck ring and actin cables. nsher012@ucr.edu
Posters for Saturday the 18th
Ashley Schwartz - Poster # 1
San Diego State
Computational Science
Graduate Student
Title: Machine learning identifies the chemical properties that predict pancreas toxicity in zebrafish
Abstract: Environmental contaminants have been linked to an increased risk of birth defects, reproductive disorders, and other adverse developmental outcomes. Researchers have turned to low-cost, high-throughput vertebrate models such as the zebrafish to study whole animal bioactivity following chemical exposures to study the etiology of disease during development. Many embryonic development transcriptomic studies, which use RNA sequencing analysis for an unbiased snapshot of gene expression following chemical exposure in the zebrafish model, have been published for the analysis of one chemical treatment. The aggregation of these publicly available datasets could yield additional information about how different chemical properties may influence differential gene expression and resulting adverse health outcomes for predictive modeling. However, advanced scientific and computational modeling techniques are needed to combine and investigate transcriptomic datasets from different research groups across the country to normalize bioinformatics pipelines. Here, we provide a case-study that supports the aggregation and analysis of transcriptomic datasets using high performance computing and unsupervised machine learning to identify key chemical properties involved in similar gene expression outcomes in the zebrafish model. Publicly available whole-embryo RNA-sequencing data were obtained for 33 environmental contaminants including 10 flame retardant chemicals, 22 polycyclic aromatic hydrocarbons, and one dioxin. We report the differential gene expression outcomes for genes involved in pancreatic development, a common organ of interest in developmental toxicity studies due to its involvement in metabolism and its relationship to the onset of diabetes. We perform unsupervised machine learning clustering techniques including hierarchical and k-means to identify the impact of clustering methodology choice on predictive outcomes. We identified specific chemical properties including chemical class, molecular weights, and bioconcentration factors that are key features driving clustering outcomes for genes involved in pancreas development. These results indicate that chemical properties and disposition of polycyclic aromatic hydrocarbons may play a role in pancreatic development. Future research will build on this methodology to develop large-scale computational models to improve the toxicological evaluation of new and emerging pollutant effects on organogenesis. avschwartz@sdsu.edu
Marycruz Flores - Poster # 2
Notre Dame
Postdoctoral Researcher
Title: Gαq homeostasis contributes to organ size regulation in Drosophila melanogaster
Abstract: G proteins mediate cell responses to a diverse set of signals. In particular, the G proteinsubunit Gαq plays pivotal role in wound healing and dysregulation of Gαq and Ca2+signaling in human causes birth defects and multiple diseases including cancer.However, the downstream effectors of Gαq are still poorly understood. Gαq regulatesdevelopmental phenotypes through Ca2+-dependent mechanisms. Ensuring Gaqhomeostasis in one organ is important for the whole organism. To investigate the role ofGαq in epithelial development, we characterized how the G protein subunit Gαq tunesthe size and shape of the Drosophila wing. Disruption of Gαq in the wing disc affectwing size and impacted the timing of pupariation. Further, Gαq upregulation wassufficient to induce organ-scale Ca2+ waves in the wing disc with a concomitantreduction in the Drosophila final wing size and pupariation delay. A subsequent RNAseqanalysis showed that disruption of Gαq homeostasis affects nuclear hormonereceptors, JAK/STAT, and Toll pathway components, among others. We identifieddifferentially expressed genes that putatively contribute to the perturbation’s phenotypefollowed by validation through immunohistochemistry. Notably, disruption of Gαqhomeostasis increased expression levels of Dilp8 signaling peptide in wing discs, a keyregulator of growth and pupariation timing, via ecdysone signaling inhibition. Overall,Gαq mediated signaling contributes to the regulation of organ size, viability, anddevelopmental timing. mflores@nd.edu
Authors: Maria F. Unger1, Vijay Velagala,1 Dharsan K. Soundarrajan1, Marycruz Flores-Flores (presenting author)1, David Gazzo1, Nilay Kumar1, Jun Li3, Jeremiah Zartman1, 2
1Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, IN 46556
2Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556
3Department of ACMS,171 Hurley Hall, University of Notre Dame, Notre Dame, IN
465562 Department of Applied and Computational Mathematics and Statistics
Dongwei Sun - Poster # 3
University of California, Riverside
Bioengineering
Postdoctoral Fellow
Title: Computational modeling of the behavior for epicardial adipose tissue derived stromal cells and left ventricular stromal cells under ischemic insults
Abstract: Stem cell therapy is a successful approach for repairing and regenerating ischemic cardiac tissues. Both epicardial adipose tissue derived stromal cells (EATDS) and left ventricular stromal cells (LVSCs) offer immense healing potential owing to the anatomic proximity, common vasculature networks and similar origin.$
EATDS and LVSCs were harvested from the heart tissues of experimental pig models and maintained under standard culture conditions. Ischemia was induced by culturing the cells using the ischemic buffer for 48 h. The cells were imaged on every 10 min intervals for 48 h against normal controls and time lapse videos were created. The alterations in the patterns and trends in cell behavior including migration, agglomeration, and division regarding assessed using the time-lapse videos.
Both CF and EAT cells prefer to agglomerate and form clustering, which speeds up the migrations and divisions. The cells under high density exhibited much more interaction and higher growth speed than medium and low density. When reaching a high cell confluency, both cells showed improved alignments when forming monolayers. Under ischemia, both cells showed reduced proliferation rates, poor alignment, and less polarized morphology.
We developed computational models based on the subcellular element method to simulate dynamics of many interacting cells. The model parameters for cells are calibrated by experimental data obtained for EATDS and LVSCs. We tested hypothesized effects of ischemic conditions on model parameters corresponding to normal control conditions. Computational results elucidates how an individual cell as well cell clusters respond to abnormal conditions. Further development of these computational models will serve to assess various stem cell therapeutic strategies. This is a joint work with Dr. Huinan Liu, Dr. Mark Alber, Dr. Jia Gou, Dr. Mykhailo Potomkin, Dr. Finosh Thankam, and Marc Christian Encarnacion. dsun027@ucr.edu
Fan Lu - Poster # 4
University of California, Santa Cruz
Applied Mathematics
Postdoctoral Fellow
Title: Accelerating Wound Healing using Deep Reinforcement Learning
Abstract: We propose a deep reinforcement learning (DRL) based approach to expedite the wound healing process. Our method efficiently divides the vision-based control task into two distinct modules: a perception module and a controller module. By separating vision-based control into a perception module and a controller module, we train a DRL agent without sophisticated mathematical modeling. Our research demonstrates the algorithm's convergence and establishes its robustness in accelerating the wound-healing process. The proposed DRL methodology showcases significant potential for expediting wound healing by effectively integrating perception, control, and predictive modeling, all while eliminating the need for intricate mathematical models. flu16@ucsc.edu
Gagan Acharya - Poster # 5
University of California, Riverside
Electrical Engineering
Graduate Student
Title: Data-driven dynamical modeling of the human brain response under deep neurostimulation
Abstract: Closed-loop (a.k.a. responsive) deep brain stimulation (DBS) has gained significant interest in the treatment of drug-resistant epilepsy (DRE) and has shown great promise for close to a decade. Due to the mechanistic complexity of DRE, however, the state-of-the-art clinical solutions for closed-loop DBS rely heavily on rigid and manual parameter tuning, while fully closed-loop seizure control algorithms have remained a theoretical possibility bottlenecked by the lack of precise computational models for the brain’s dynamic network response to DBS. In our work, we pursue the problem of data-driven learning of neurodynamical models to accurately predict the large-scale network evoked in the brain due to stimulation. To this end, we compare a family of linear and non-linear models based on their efficacy in predicting the observed iEEG response to stimulation input of varying current amplitudes and frequencies. Our analysis of DBS response data from multiple subjects and brain locations indicates that the evoked activity can best be explained using stimulation-triggered switched linear models. We find that 250ms of historical data is sufficient to model the evolution of iEEG voltage dynamics, with longer histories leading to only marginal improvements in the overall prediction accuracy. A key observation is that model comparisons are sensitive to the criteria used such as mean squared error (MSE) and Wilcoxon signed-rank test with each method leading to different versions of the ‘best’ model. We observe great heterogeneity in the best models of non-stimulated channels when we use the latter method, while MSE almost uniformly favors the switched linear model with exogenous input. Despite this mismatch, we observe consistent modeling patterns across subjects that are agnostic to the criterion used. Among them is the importance of network interactions and historical lags of electrode channels in substantially decreasing the validation error. Also, our preliminary results indicate that the nature of interactions seen in the ‘best’ model is correlated to the precise xyz coordinates (but not the mere brain region) of the stimulation site. We observe that combining training data acquired with multiple stimulation parameters (frequency, for example,) is beneficial in decreasing the residual error. This includes a significant benefit from including abundantly-available resting state data in training stimulation models, further highlighting the significance of experimental design in the system identification of brain dynamics. gacha002@ucr.edu
Kim Ngan (Luna) Huynh - Poster # 6
San Diego State
Computational Science Research Center
Graduate Student
Title: The Associations Between Sleep Quality, Physical Activity and Pregnancy Outcome
Abstract: Majority of women experience poor sleep quality as their body changes during pregnancy. Poor sleep quality during pregnancy may increase the risk for pregnancy complications. Physical activity may improve sleep quality in pregnant women. The aim of this study was to build a novel approach to examine if sleep quality and physical activity during week 22 of gestation are associated with pregnancy outcome. The rest-activity and health data for 43 pregnant women were obtained from a longitudinal observational prospective cohort study. The women’s rest-activity were measured at gestational week 22 using a wrist actigraph device for seven consecutive days. We assigned a woman to a complicated pregnancy group if her newborn was premature, postmature, had abnormal birth weight, abnormal birth length, disease, prolonged stay at the hospital after birth or if the mother was diagnosed with preeclampsia, hypertension and/or pregestational diabetes. The women’s sleep quality was determined by calculating the variability in rest activity during sleep time for six consecutive days. The variability in rest activity was quantified using the Wasserstein distance, a metric often used in optimal transport theory. The intensity of physical activity was determined by calculating the area under the curve (AUC) of the rest activity plots. This study found that women with uncomplicated pregnancy were more likely to have better sleep quality and were more active than women in the complicated pregnancy group in gestational week 22. This study has introduced a novel approach to analyze the rest activity of pregnant women and the findings adds to the body of research that emphasize the impact of sleep quality and physical activity on pregnancy. lhuynh2785@sdsu.edu
Kyle Nguyen - Poster # 7
North Carolina State University
Biomathematics
Graduate Student
Title: Quantifying fluidization patterns in mesenchymal cell populations using topological data analysis and agent-based modeling
Abstract: Fibroblasts in a confluent monolayer are known to adopt elongated morphologies in which cells are oriented parallel to their neighbors. We collected and analyzed new microscopy movies to show that confluent fibroblasts are motile and that neighboring cells often move in anti-parallel directions in a collective motion phenomenon we refer to as ``fluidization’’ of the cell population. We used machine learning to perform cell tracking for each movie and then leveraged topological data analysis (TDA) to show that time-varying point-clouds generated by the tracks contain significant topological information content that is driven by fluidization, i.e., the anti-parallel movement of individual neighboring cells and neighboring groups of cells over long distances. We then utilized the TDA summaries extracted from each movie to perform Bayesian parameter estimation for the D’Orsgona model, an agent-based model (ABM) known to produce a wide array of different patterns, including patterns that are qualitatively similar to fluidization. Although the D’Orsgona ABM is a phenomenological model that only describes inter-cellular attraction and repulsion, the estimated region of D’Orsogna model parameter space was consistent across all movies, suggesting that a specific level of inter-cellular repulsion force at close range may be a mechanism that helps drive fluidization patterns in confluent mesenchymal cell populations. kcnguye2@ncsu.edu
Manasa Kesapragada - Poster # 8
University of California, Santa Cruz
Applied Mathematics
Graduate Student
Title: Data-driven classification of cell subtypes based on time-lapse microscopy of single cells
Abstract: Single-cell time-lapse microscopy allows researchers to track the dynamic response of cellular processes in real time. Observing these dynamics to external stimuli can help researchers understand the regulation mechanisms underlying complex biological processes. When studying the evolution of cell subtypes, it can become difficult to identify the cell types. Cell size and shape have been used to characterize these cell subtypes, but quality images that can provide cell morphology are difficult to come by. We propose that motility properties can be mapped to cell morphology and, hence, cell subtype. We applied this to macrophages, critical players in our body's defense and wound healing. We found that different types of macrophages, move uniquely. We developed methods to track single cells and linked cell movement to its shape using machine learning. Through this study, we demonstrate that mapping migratory patterns and motility properties to cell morphology can inform the classification of cell subtypes. mkesapra@ucsc.edu
Marc Encarnacion - Poster # 9
University of California, Riverside
Department of Computer Science
Undergraduate Student
Title: Computational model for cell migration of epicardial adipose tissue derived stromal cells
Abstract: Epicardial adipose tissue derived stromal cells (EATDS) is a useful tool for regenerating damaged heart tissue. To find conditions optimal for tissue development and propose efficient therapeutic strategies for inflicted tissues, it is important to study the dynamics of many interacting such cells. In particular, we are interested in trajectories and morphology characteristics of EATDS cells as they proliferate and form a connective tissue. Using experimental data from Dr. Liu’s group from University of California, Riverside and Dr. Thankam’s lab from Western University of Health Sciences, we develop an agent-based detailed model for EATDS cells. This model captures various shapes of individual cells, their motility, cell proliferation, both steric and adhesive interactions, as well as cell-cell alignment. Specific rules and range of model parameters are calibrated using experimental data. The objective of the model is to suggest how one can control confluency rate and topology of the resulting tissue by changing initial conditions. This is joint work with Huinan Liu, Mark Alber, Finosh Thankam, Mykhailo Potomkin, Jia Gou, and Dongwei Sun. My work is funded by the California Institute of Regenerative Medicine. menca003@ucr.edu
Parsa Ghadermazi - Poster # 10
Colorado State University
Chemical and Biological Engineering
Graduate Student
Title: Microbiome Evolution from a New Perspective: Reinforcement Learning Provides an Evolutionary Perspective of Microbial Interactions.
Abstract: Microorganisms are integral to ecosystems, shaping their environment and material flow by forming a complex network of interacting cells. Predicting the phenotype of microorganisms from their genotype has motivated creation of mathematical models to describe the behavior of microbial communities. While metabolic modeling methods based on flux balance analysis (FBA) offer insights into homogenous and heterogenous microbial community metabolism, it falls short in predicting long-term stability, especially in presence of interacting microbes. We propose "Self-Playing Microbes in Dynamic FBA", a novel reinforcement learning algorithm. It treats microbial metabolism as a decision-making process, enabling microorganisms to adapt metabolic strategies for enhanced fitness in a dynamic context by trial and error in a simulation environment and finding flux regulation policies that stabilize in a microbiome in the presence of other microbes, by relying on first principles of microbial ecology with minimal reliance on pre-determined strategies and experimental observations. Our work demonstrates improved performance over existing methods in various scenarios, such as, metabolite exchange between auxotrophs and secretion of extracellular enzymes by the cells highlighting its biological significance parsa96@colostate.edu
Robert McDonald - Poster # 11
Harvard University/University of Oxford
Engineering and Applied Sciences
Mathematics
Graduate Student
Title: Zigzag persistence for coral reef resilience using a stochastic spatial model : A complex interplay between species governs the evolution of spatial patterns in ecology.
Abstract: An open problem in the biological sciences is characterizing spatio-temporal data and understanding how changes at the local scale affect global dynamics/behaviour. Here, we extend a well-studied temporal mathematical model of coral reef dynamics to include stochastic and spatial interactions and generate data to study different ecological scenarios. We present descriptors to characterize patterns in heterogeneous spatio-temporal data surpassing spatially averaged measures. We apply these descriptors to simulated coral data and demonstrate the utility of two topological data analysis techniques—persistent homology and zigzag persistence—for characterizing mechanisms of reef resilience. We show that the introduction of local competition between species leads to the appearance of coral clusters in the reef. We use our analyses to distinguish temporal dynamics stemming from different initial configurations of coral, showing that the neighbourhood composition of coral sites determines their long-term survival. Using zigzag persistence, we determine which spatial configurations protect coral from extinction in different environments. Finally, we apply this toolkit of multi-scale methods to empirical coral reef data, which distinguish spatio-temporal reef dynamics in different locations, and demonstrate the applicability to a range of datasets. robertmcdonald@fas.harvard.edu
Sandor Volkan-Kacso - Poster # 12
Caltech
Chemistry and Chemical Engineering
Research Scientist
Title: Extracting short-lived states from single-molecule trajectories in rotary molecular machines
Abstract: Single-molecule imaging has the potential to reveal the multi-timescale behavior of nanoscale machines. One example is the rotation of the F1-ATPase enzyme in which a substep can have microsecond transition dynamics yet waiting times vary from milliseconds to seconds. Recently, we proposed a method to analyze fast (10-100µs timestep) rotation trajectories in F1-ATPase using the distribution of jumps in the rotation angle. The analysis involves the transitions during the stepping between subsequent catalytic dwells. A theoretical-computational approach based on a multi-state model is used to model fluctuation of the imaging probe as the molecular motor undergoes stepping rotation. A key quantity in this method is the angular velocity vs. rotation angle extracted from both experimental data and computer simulations. When applying the method on Thermophilic Bacillus F1-ATPase rotation data, we detected the presence of a short-lived substep previously not detectable in the histograms. The comparison between the experimental and theory reveals that an 80O substep of the “concerted” ATP binding and ADP release involves an intermediate state reminiscent of a 3-occupancy structure. Its lifetime (~10 µs) is about six orders of magnitude smaller than the lifetime for “spontaneous” ADP release from a singly occupied state. The ~10 µs lifetime is comparable with the experimental imaging frame time, so by detecting this short-lived state the method provides an avenue towards temporal super-resolution. Most recently, this method was applied to single-molecule imaging data from Paracoccus Denitrificans F1-ATPase and it yielded a similar hidden state in the transitions between subsequent long dwells. Our recent findings indicate a common mechanism for the acceleration of ADP release in the F1-ATPase motor of the two species. svk@caltech.edu
References
N. Suiter & S. Volkán-Kacsó, Frontiers in Mol. Biosci. 10, 1184249 (2023)
S. Volkán-Kacsó and R. A. Marcus, Frontiers in Microbiol, 13, 861855 (2022)
S. Volkán-Kacsó, et al., Proc. Natl. Acad. Sci., USA, 116, 25456 (2019)"
Sashiel Vagus - Poster # 13
San Diego State
Mathematics
Graduate Student
Title: Quantifying entropy values for pregnant women's rest and activity patterns during their gestational period in order to dictate a good versus bad pregnancy
Abstract: Recent studies suggest that there is a link between rest-activity patterns during pregnancy and maternal-fetal health. Studies also suggest that irregular sleep patterns can negatively affect metabolic and hormonal physiology. More research is needed to fully understand how women's rest-activity patterns affect maternal-fetal health. This study proposes a method to relate women's rest-activity during pregnancy to maternal and fetal health outcomes. Rest-activity curves were obtained from a previous study on a group of pregnant women in which their rest activity during gestational week 22 (G22; n=41) and gestational week 32 (G32; n=44) were monitored. After we obtained the rest-activity curves, the daily and weekly activity patterns for each woman was analyzed using persistent homology and entropy. The vietoris-rips complex filtration was applied to each woman's activity curve to obtain their persistence diagrams. The entropies of the persistence diagrams were quantified. Results showed that the women who had a higher variability in their daily entropy values during gestational week 22 were more likely to have a bad pregnancy. In particular, women with an entropy value above 8.25 during gestational week 22 were more likely to have a bad pregnancy. Our study found that (1) the degree of variability in the women's entropy values was negatively associated with the women's body mass index (BMI) during gestational week 22 and week 32 and (2) the degree of variability in the women's entropy values was positively associated with the women's blood glucose. Our findings support the idea that persistence homology and entropy may be used to relate activity rhythms during pregnancy to certain unfavorable health outcomes for the mother and fetus. sashiel.vagus@gmail.com
Shannon McFadden - Poster # 14
Huntington Beach High School / UCI
Andrew Rusli
St. Margaret’s Episcopal School / UCI
Title: Astral numbers and the mechanical strength of cells
Abstract: Cells have the ability to control their rigidity - that is, the structural property of a material that allows it to withstand mechanical force - over orders of magnitude in minutes. This is an ability that material engineers aspire to recreate. It is also an ability that goes wrong in many diseases in which cells are either too stiff (parasitic infections like malaria, some anemias) or too soft (some cancers and heart disease). Recent work has shown that part of the cell's rigid structures (F-actin) into star-shaped patterns called asters that then crosslink into larger networks. In this work, we show that there is an optimal number of filaments per aster -- what we term the "astral number" -- that maximizes rigidity. This work provides a direct connection between a readily-measurable quantity (astral number) to a biomedically-relevant but harder-to-measure one (cell rigidity). This is a joint work with Brady Berg and Jun Allard.
Ta I Hung - Poster # 15
UC Riverside
Chemistry
Graduate Student
Title: Residue Correlation Network Guided Protein Engineering to Enhance Protein-Protein Interactions
Abstract: Designing protein binders that are both stable and specific to target a particular protein is a difficult and time-consuming task in protein engineering. This poster introduces a new strategy for efficient protein engineering. Using molecular modeling, we revealed a residue interaction and dihedral angle correlation network of a protein-protein complex to guide the selection of a minimal number of mutated residues on a protein surface to create highly potent and selective protein-based inhibitors. We demonstrated our strategy using ubiquitin (Ub) and MERS coronaviral papain-like protease (PLpro) complexes, where Ub is one central player in many cellular functions and PLpro is an antiviral drug target. By utilizing a combination of molecular dynamics simulations and experimental assays, we were able to predict and confirm the binding of our designed Ub variant (UbV) binders. Our designed UbV with 3 mutated residues at the hydrophobic core and the Zn binding region, resulted in a ~3,500-fold increase in functional inhibition, compared with the wild-type Ub. Further optimization using Ub C-terminal created a 5-point mutant that achieved a KD of 1.5 nM and IC50 of 9.7 nM. The modification led to a 27,500-fold and 5,500-fold enhancements in affinity and potency, respectively, as well as improved selectivity, without destabilizing the UbV structure. Introducing minimal mutation site our design UbV remain high thermal stability. Our approach effectively designs protein binders with higher binding affinity, which assists the development of therapeutics for deubiquitinase systems and cell biology studies to investigate specific Ub signaling regulation and protein-protein interaction. thung002@ucr.edu
Timmy Liang - Poster # 16
SDSU
Applied Mathematics
Graduate Student
Title: Modeling HIV Latent Infection Under Drugs of Abuse
Abstract: Despite the tremendous success of antiretroviral therapy (ART), there is no cure for HIV due to the establishment of latently infected cells. While the drugs of abuse, such as opiates, have been shown to increase viral load and pathogenesis, how drugs of abuse affect the HIV latent infection is not wellunderstood. In this study, we develop a mathematical model to investigate how the drugs of abuse can affect latent infection dynamics. The model is validated using experimental data from HIV infection of humans and Simian Immunodeficiency Virus (SIV) infection of morphine-addicted macaques. Our model shows that the dynamics of latently infected cells can be significantly altered due to the presence of the drugs of abuse." tliang7106@sdsu.edu
William Troxel - Poster # 17
University of California, Riverside
Chemistry
Graduate Student
Title: Surveying Imatinib:Kinase Affinities and Specificities Using Molecular Dynamics Simulations
Abstract: Computational chemistry allows us to model drug-protein interactions, structural changes, and functions, which deepens our understanding in molecular recognition. Most drugs specifically target one protein, but some interact with multiple non-targeted proteins. Imatinib binds to ABL kinase and inhibits its inactive state to treat chronic myeloid leukemia. Protein assays show it interacts with other kinases to treat other tumors, but the atomistic mechanisms are not well-understood. Complex structures are available, but they do not show binding dynamics or mechanisms. Our group has been carrying out multiple 100 ns to 500 ns-length molecular dynamics (MD) simulations. This poster reports my analysis of the MD trajectories to categorize the important bound-state interactions dictating imatinib’s kinase affinities and specificities. This poster will focus on kinases ABL, KIT, LCK, p38, and SRC. The kinase-imatinib complex MD runs are all stabilized after 55 ns based on the root-mean-square-deviation. The root-mean-square-fluctuation for ABL, LCK, p38, and SRC shows high flexibility around residues 150-170, corresponding with the alpha-c-helix, a critical kinase regulatory motif. KIT does not show significant flexibility in the bound state at the same region, which is expected from previous literature. The poster will also discuss results from molecular docking, hydrogen bond analysis, and pairwise force distribution analysis between the protein and imatinib. Our study also shows that imatinib’s multi-kinase promiscuity depends on more than protein sequence identity and homology to ABL. A closer study of the non-covalent interactions is necessary to fully understand imatinib’s protein promiscuity. wtrox002@ucr.edu
Xinru Qiu - Poster # 18
University of California, Riverside
Biological Sciences
Postdoctoral Fellow
Title: Transcriptional heterogeneity of activated platelets correlates with negative outcomes in sepsis and COVID19 and plays a role in autoimmune diseases
Abstract: Single-cell transcriptomic profiling of peripheral blood mononuclear cells (PBMCs) in patients with COVID-19, sepsis, and systemic lupus erythematosus (SLE) has revealed insights into disease mechanisms. We hypothesize that the increased platelet population in PBMC fractions in severe cases represents activated platelet subpopulations responsible for disease outcomes in acute inflammation-driven diseases, suggesting potential new therapeutic strategies targeting these abnormal platelet subtypes. By collecting and integrating scRNAseq data from publicly available datasets on COVID-19, sepsis, and SLE, and using SingleR, Seurat, and Monocle analysis software, unique platelet subpopulations were identified. These subpopulations were found to correlate with disease severity and outcomes. Dynamic analysis revealed how these platelets behave and function under various conditions. Abnormal platelet subpopulations were found to overexpress genes related to endotheliopathy, potentially increasing the risk of disseminated intravascular coagulation in fatal patients, as well as genes modulating lymphocyte function, suggesting a broader role for platelets in abnormal inflammatory and immune responses. xqiu014@ucr.edu
Yuan Xu - Poster # 19
UCSC
Genomics Institute and Biomedical Engineering
Graduate Student
Title: Determination of the centromere protein A (CENP-A) landscape at single-molecule resolution
Abstract: Centromere protein A (CENP-A) is a histone H3 variant that specifies the location of each chromosome’s centromere, which is essential for proper kinetochore attachment and chromosome segregation. Mislocalization and misregulation of CENP-A can drive chromosomal breakage and rearrangement, leading to cancer and chromosomal aneuploidies. Measuring and comparing the density and spacing of CENP-A-containing nucleosomes within and between endogenous human centromeres will provide new insights into the structure of the inner kinetochore, as well as how this structure helps to ensure proper centromeric function and strength. However, mapping the locations of CENP-A nucleosomes within centromeric regions remains challenging due to their repetitive nature. Sequencing approaches that have been developed to map protein-DNA interactions have in the past relied on short-read sequencing data (e.g. NChIP, XChIP, CUT&RUN, CUT&Tag). However, assigning short reads to unique positions within highly repetitive centromeric regions remains difficult or impossible. Recently, directed methylation with long-read sequencing (DiMeLo-seq) was developed, which deposits exogenous adenine methylation marks near a desired protein, then uses nanopore long-read sequencing to quantify this exogenous adenine methylation directly along with endogenous cytosine methylation. To study protein-DNA interactions in the highly repetitive centromeric regions of the human genome, this approach overcomes the limitations of short-read sequencing in the ability to observe multiple binding events on long, single DNA molecules that are mappable to repetitive regions. Here, we present the distribution of CENP-A on ultra-long single molecules by using DiMeLo-seq on the HG002 cell line. Furthermore, we discuss a probabilistic algorithm for predicting the positions of single CENP-A nucleosomes on single chromatin fibers. yxu267@ucsc.edu
Yutong Sha - Poster # 20
UC Irvine
Math
Postdoctoral Fellow
Title: Reconstructing transition dynamics from static single-cell genomic data
Abstract: Recently, single-cell transcriptomics has provided a powerful approach to investigate cellular properties in unprecedented resolution. However, given a small number of temporal snapshots of single-cell transcriptomics, how to connect them to obtain their collective dynamical information remains an unexplored area. One major challenge to connecting temporal snapshots is that cells measured at one temporal point may divide at the next temporal point, leading to growth and differentiation in the system. It’s increasingly clear that without incorporating cellular growth dynamics, the inferred dynamics often becomes incomplete and less accurate. To fill these gaps, we present a novel method to reconstruct the growth and dynamic trajectory simultaneously as well as the underlying gene regulatory networks. A deep learning-based dynamic unbalanced optimal transport is developed to infer interpretable dynamics from high-dimensional datasets. ysha3@uci.edu
Babgen Manookian - Poster # 21
City of Hope
Department of Computational and Quantitative Medicine
Postdoctoral Fellow
Title: Bayesian Model Study on Temporal Dependency of Residue Pairs in Class A GPCR Proteins: Insights into Allosteric Mechanisms
Abstract: Protein dynamics are pivotal in the function and activity of Class A GPCR proteins. Understanding the underlying function mechanisms is of paramount importance for drug discovery and the development of targeted therapeutics. We hypothesize that the interplay between residues involved in GPCR activity have distinct temporal dependencies that, if uncovered, will provide insight on the protein function mechanism. Dynamics Bayesian models were leveraged to investigate temporal dependencies between residue pair contacts in the protein. The analysis was conducted using molecular dynamics trajectory data of class A GPCRs undergoing conformational changes. We incorporated: (1) the rescoring of static Bayesian network using subsets of the data along the trajectory, and (2) the embedding of the most relevant lag variables via conditional mutual information. We demonstrate that interaction between contact pairs vary throughout the molecular dynamics trajectory. Specifically, there exists a silence in all dependencies immediately following transition. We identified contact pairs with increased dependency are located in critical regions of the protein known to play a role in the activity. This research project deepens our comprehension of Class A GPCR protein dynamics by inferring on the mechanism of protein activity by determining the temporal dependencies between residues. bmanookian@coh.org
Zirui Zhang - Poster # 22
UC Irvine
Mathematics
Postdoctoral Fellow
Title: Parameter Inference in Diffusion-Reaction Models of Glioblastoma Using Physics-Informed Neural Networks
Abstract: Glioblastoma is an aggressive brain tumor that proliferates and infiltrates into the surrounding normal brain tissue. The growth of Glioblastoma is commonly modeled mathematically by diffusion-reaction type partial differential equations (PDEs). These models can be used to predict tumor progression and guide treatment decisions for individual patients. However, this requires parameters and brain anatomies that are patient specific. Inferring patient specific biophysical parameters from medical scans is a very challenging inverse modeling problem because of the lack of temporal data, the complexity of the brain geometry and the need to perform the inference rapidly in order to limit the time between imaging and diagnosis. Physics-informed neural networks (PINNs) have emerged as a new method to solve PDE parameter inference problems efficiently. PINNs embed both the data the PDE into the loss function of the neural networks by automatic differentiation, thus seamlessly integrating the data and the PDE. In this work, we use PINNs to solve the diffusion-reaction PDE model of glioblastoma and infer biophysical parameters from numerical data. The complex brain geometry is handled by the diffuse domain method. We demonstrate the efficiency, accuracy and robustness of our approach.ziruz16@uci.edu
Lingxia Qiao - Poster # 23
University of California, San Diego
Postdoc
Title: Dynamics of Calcium-cAMP oscillations as a function of membrane nanodomains of AKAP-VGCC-PKA clusters
Abstract: The nanoscale organization of key enzymes is critical for the compartmentation of cyclic adenosine monophosphate (cAMP) and encodes various cellular processes. Previous experimental studies have shown the formation of nanoclusters of A-kinase anchoring protein 79/150 (AKAP150) and adenylyl cyclase 8 (AC8) at the surface of pancreatic MIN6 beta cells. These clusters regulate cAMP-Ca^2+ oscillatory phases, in which AC8 modulates cAMP oscillations to become in-phase with Ca^2+ oscillations. In this work, by using a computational model, we investigated how cAMP and Ca^2+ oscillations are affected by biophysical aspects of cells and the distribution of AKAP150/AC8 nanoclusters. We found that different membrane curvatures have little impact on the phase delay between cAMP and Ca^2+ oscillations. Nevertheless, the surface-to-volume ratio of cells shows a big impact on the phase delay: the smaller the surface-to-volume is, the longer the time delay is. In addition, the distribution of AKAP150/AC8 nanoclusters also plays an important role in regulating phase delay, where the increased number of AKAP150/AC8 nanoclusters leads to the decreased time delay between cAMP and Ca^2+ oscillations. Furthermore, we used STROM data as the distribution of AKAP150/AC8 nanoclusters and showed the effects of membrane curvature and surface-to-volume ratio on the time delay, which are consistent with previous simulations. These findings reveal the factors that may affect the phase delay between cAMP and Ca^2+ oscillations. liqiao@ucsd.edu
Asees Kaur - Poster # 24
University of California, Merced
Title: Improving DSA Image Segmentation with CNNs
Abstract: This poster introduces an innovative method for the segmentation of Digital Subtraction Angiography (DSA) images, a critical task in medical image analysis. Using Convolutional Neural Networks (CNNs), our approach employs an iterative process to expand segmented regions in growing directions. A CNN predicts class probability scores within a small pixel neighborhood, determining pixel inclusion based on a threshold. The process continues iteratively until no new pixels qualify for inclusion. Our method achieves remarkable segmentation accuracy while preserving biological features. akaur101@ucmerced.edu